In my previous post, I described the summary report that accompanied my 23andMe exome sequence data, with summary statistics and the filtering scheme used to arrive at a list of 21 rare, moderate-to-high predicted impact variants. Here I will show how I used publicly available, free and open-source tools to explore my exome sequence further. I started with the 23andMe list of 21 variants of interest. My questions were:
- Can I visualize these variants in the mapped sequence reads? See trust but verify below.
- What are the functions of the affected genes?
- How likely are the variants to seriously affect the functions of their genes?
Answers to these questions would allow me a better sense of how interesting or worrisome these variants may be, not just for my health, but for my daughter, who has a 50% chance of inheriting any one of these variants.
Bam files and samtools
The bam file is a compressed, binary version of the SAM (sequence alignment/map) file format. This file contains all of the sequence reads aligned to the reference genome sequence. These sequence alignments are used to determine whether your DNA has a sequence difference from the reference, whether you are heterozygous or homozygous for the variant, and how statistically reliable the genotype determination is, based on the number of times that particular base has been read (depth of coverage) and the quality of the sequence reads at that position. This alignment information (without the actual sequences) is captured in the index file, with a .bai filename extension.
If you are comfortable working with the command line, samtools is a software package that allows you to work with the bam file, to extract alignments for individual chromosomes, to convert bam to sam and vice versa, to create .bai index files, and to export the variant calls in the .vcf variant call format.
One way to actually see the reads aligned against the reference is to use the Integrative Genomics Viewer (IGV) from the Broad Institute. You select the reference genome used to create the alignments in the bam file, and load your bam file. The genome viewer uses the information in the .bai index file to show all the sequence reads ?piled up? below the reference sequence. You can load multiple bam files (each must have a corresponding .bai index file) for comparison.
Example: my CF allele
I was somewhat surprised that a CFTR variant showed up on my 23andMe report. I use cystic fibrosis to integrate multiple concepts in my Intro Biology class (see my post on CF, a case study for membranes and transport). I knew that CF was most common among Northern Europeans, with much lower frequencies in Asians. But here it was, a mutation causing a non-conservative amino acid substitution. Changing glutamic acid (E) to glycine (G) could have serious consequences for either protein folding or activity, because these two amino acids are unalike.
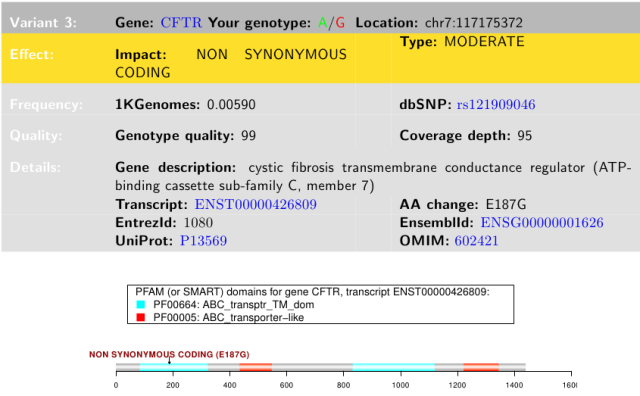
23andMe report for my CFTR variant
Integrative Genome Viewer
To see the underlying data for myself, I loaded up my bam file in the IGV:
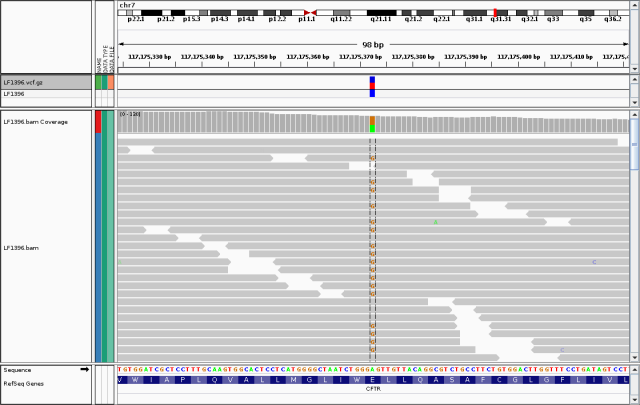
Snapshot of IGV window showing reads aligned to chr7:117175372. At this position, approximately half the reads in LF1396.bam have the reference ?A? and half have the alternate ?G?. The track above shows LF1396.vcf.gz (the gzipped version of the vcf file).
My heterozygous genotype for this variant is obvious, and well-supported by high read depth and almost equal numbers of reads bearing the two alleles.
NCBI and dbSNP
This variant is identified as a previously characterized single nucleotide polymorphism (SNP) in dbSNP. I looked it up by going to the NCBI home page and simply entering ?rs121909046? in the search field and selecting the SNP database. The dbSNP page provides a wealth of information about this particular variant, identifying it as a probable pathogenic variant.
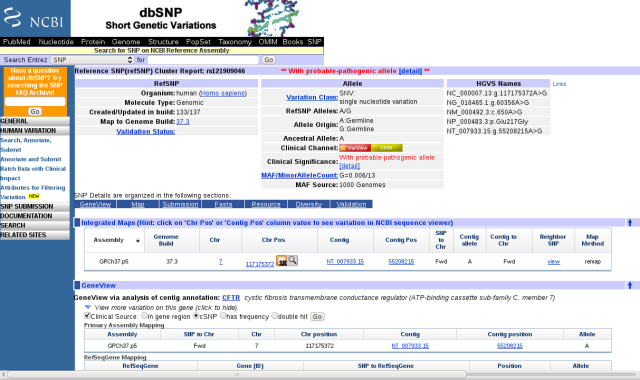
dbSNP page for rs121909046
Before I got too excited about the pathogenic part, I noted that this SNP is annotated as Glu217Gly, or E217G, affecting the 217th amino acid in the CFTR protein, whereas the 23andMe report annotates this as E187G, affecting the 187th amino acid. A quick check of the human CFTR protein sequence showed that the 187th amino acid is Asn (= N in single letter amino acid code). I don?t know why the 23andMe report has this discrepancy, but similar errors in identifying amino acid changes showed up in other gene variant reports (see Trust but verify, below).
OMIM
Scrolling down, near the bottom of the dbSNP page is a link to an OMIM (Online Mendelian Inheritance in Man) entry. Clicking on it took me to the subsection of the OMIM article on CFTR that cites a journal article concerning this variant. I followed links to the article by Lee et al. (2003), which is available free full-text. I learned that the E217G variant appears in the Korean population with an allele frequency around 1.3%, and that heterozygotes have a higher risk of bronchiectasis, but not of pancreatic insufficiency. Molecular studies revealed that this mutation causes 60% reduction in the amount of CFTR protein that appears on the membrane. It is a relatively mild disease allele.
UCSC Genome Browser
What more can I learn about this mutation? I can view it in the UCSC genome browser, which offers some additional information. I can even load my bam or vcf file as a custom track. I decided to load my vcf file to view just the variant annotations, rather than clutter up my view with all the read alignments. To do this, I had only to go to the directory containing my exome data file, which includes the gzipped vcf file, LF1396.vcf.gz, and run the following command: tabix -p vcf my.vcf.gz? to get a binary index file LF1396.vcf.gz.tbi. I followed the helpful directions at http://genome.ucsc.edu/goldenPath/help/vcf.html to load this as a custom track.
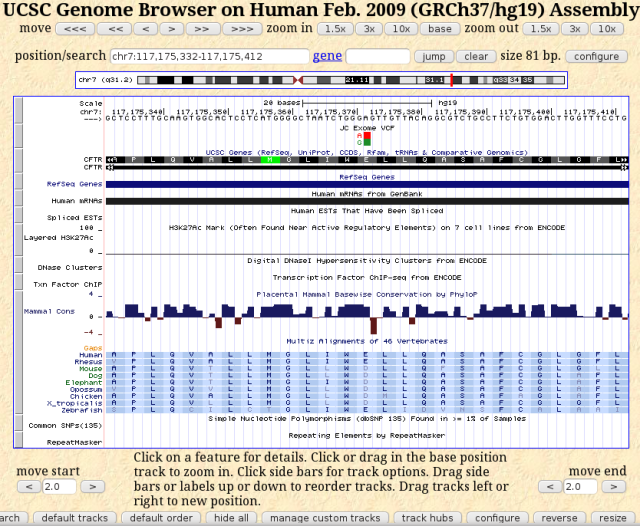
UCSC Genome Browser view of CFTR E217 gene location with JC exome added as a custom track
What I like about this view are the alignments with other vertebrates. The E217 in human CFTR is conserved throughout vertebrates, from fish to primates, with either glutamic acid (E) or aspartic acid (D) at this position. Aspartic acid and glutamic acid are chemically similar, both having acidic side chains, and are often interchangeable. Given such conservation at this position, having a glycine at this position most likely would affect the protein adversely.
Trust but verify
I used these tools to examine all 21 of my ?interesting? variants identified in my 23andMe exome summary report. Of most concern to me were variants that the report stated would cause non-conservative amino acid changes in my MSH2 and PRNP proteins.
MSH2 is a DNA repair gene. Even heterozygotes have a markedly higher risk of cancer, particularly colon cancer, because of the real possibility that a somatic mutation will knock out the only functional copy of this gene. Such cells, unable to repair DNA damage, would rapidly accumulate other mutations that could lead to cancer.
PRNP is the prion protein. Altered folding of this brain protein leads to an infectious protein particle that causes slow neurodegenerative disease, as in mad cow disease and scrapie. Mutations in this protein are associated with familial (inherited) neurodegenrative diseases.
Both my MSH2 and PRNP mutations are already in dbSNP. I learned that my PRNP variant is known to be non-pathogenic. A huge relief! But whereas the 23andMe report identified the amino acid change as E158K (glutamic acid at position 158 changed to lysine), both dbSNP and IGV show that it is actually E219K. So the amino acid change was correctly identified, but at the wrong position, just like my CFTR mutation. More puzzling, and of greater concern, was that my MSH2 mutation was identified as A43D. But dbSNP and the BAM alignment viewed in IGV both show that this is a ?silent? mutation that results in no amino acid change, involving glycine at either the 157th position or the 91st position, depending on the form of the protein (MSH2 has multiple forms resulting from alternative splicing).
I found a similar discrepancy in at least one other gene, where a silent mutation was incorrectly identified as an amino acid change. Overall, the 23andMe annotations of amino acid changes were a very mixed bag, where about half of the changes were incorrectly identified by position, or more seriously, by the type (amino acid change versus silent). This indicates a need to re-run that part of the analysis (SNPeff).
Learning my genetic heritage
Leaving aside these discrepancies on the effects of mutations, I found that one of the accurately annotated variants, in my GALK1 (galactokinase) gene, has a name, the ?Osaka variant?, and occurs with a frequency of 4% among Japanese and 3% among Koreans (Okano et al., 2001). The Osaka variant is associated with bilateral cataract formation in elderly Japanese people. My mother had to undergo Lasik eye surgery for cataracts in both eyes; I strongly suspect that I inherited this allele from her.
Galactokinase deficiency is a finding of a kind that gladdens the hearts of personal genomics practitioners, because it is an ?actionable? variant, meaning that a person can do something about it. In this case, the action is as simple as avoiding foods that contain high levels of galactose: milk and beets. I?m not fond of beets, and I drink milk only occasionally, but I would have a tough time giving up cheese and ice cream entirely.
In this exercise, I used several free, publicly available tools and databases to view and obtain a wealth of information about specific candidate variants. I learned that I carry a couple of unsuspected pathogenic alleles, for cystic fibrosis and galactokinase deficiency. Both alleles occur at relatively high frequencies in the Korean population. I don?t expect that the CF allele will affect my life, but I will be avoiding milk and moderating my ice cream consumption, in case my heterozygosity for the Osaka allele elevates my future risk for cataracts.
This is just the beginning of my exploration of my exome. Future blog posts will discuss various alternative ways to sift for ?interesting? variants.
References:
Lee, J. H., Choi, J. H., Namkung, W., Hanrahan, J. W., Chang, J., Song, S. Y., Park, S. W., Kim, D. S., Yoon, J.-H., Suh, Y., Jang, I.-J., Nam, J. H., Kim, S. J., Cho, M.-O., Lee, J.-E., Kim, K. H., Lee, M. G. A haplotype-based molecular analysis of CFTR mutations associated with respiratory and pancreatic diseases. Hum. Molec. Genet. 12: 2321-2332, 2003. [PubMed: 12952861] [Full Text: HighWire Press, Pubget]
Okano, Y., Asada, M., Fujimoto, A., Ohtake, A., Murayama, K., Hsiao, K.-J., Choeh, K., Yang, Y., Cao, Q., Reichardt, J. K. V., Niihira, S., Imamura, T., Yamano, T. A genetic factor for age-related cataract: identification and characterization of a novel galactokinase variant, ?Osaka,? in Asians. Am. J. Hum. Genet. 68: 1036-1042, 2001. [PubMed: 11231902, related citations] [Full Text: Elsevier Science, Pubget]
Like this:
Be the first to like this.
erika van pelt pat robertson hunger games trailer hunger games trailer in plain sight hunger games movie review bats
No comments:
Post a Comment
Note: Only a member of this blog may post a comment.